CORE FACILITY
Mass Spectrometry
Head: Paolo Grumati
Phospho-proteomic Coordinator: Francesca Sacco
Staff: Michele Santoro, Giuseppe Di Tullio, Lucia Santorelli, Giorgia Massacci

MS- full proteome
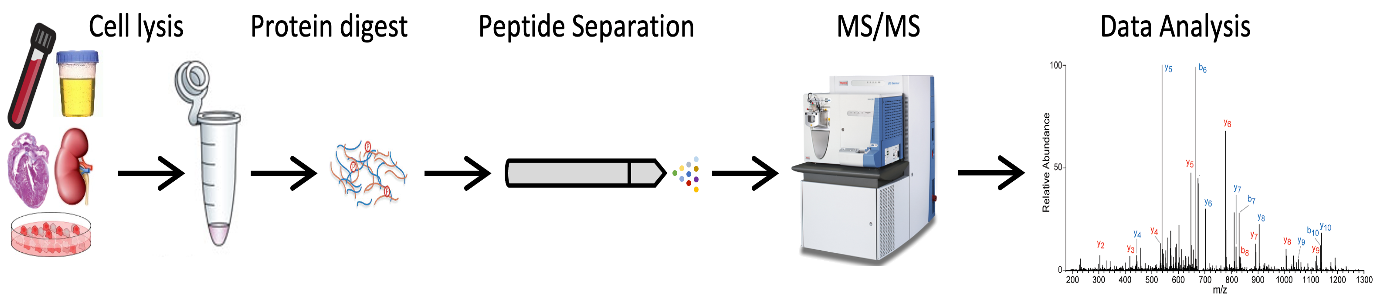
Proteins are the bricks that make biology. Abnormal proteins are often the cause of diseases, thus by targeting proteins we could generate novel cures. Full proteome analysis provides a picture of the quantity and quality of proteins present in a biological sample. Proteomics relies mainly on the “bottom-up” approach. Proteins can be extracted from several biological samples and enzymatically digested. The obtained peptides are ionized, and ions separated according to their mass/charge ratio (m/z) thus finally detected and analyzed.
MS-based full-proteome workflow
MS-based Phosphoproteomics
Protein phosphorylation modulates many biological processes. However, the elucidation of the complex regulatory circuits controlling cell responses to external and internal stimuli is still limited by our inability to describe the global phosphorylation network by systemic approaches. We set up MS-based phosphoproteomic workflow, which allows tens of thousands of phosphorylation sites to be monitored in multiple conditions.
MS-based phospho-proteomics workflow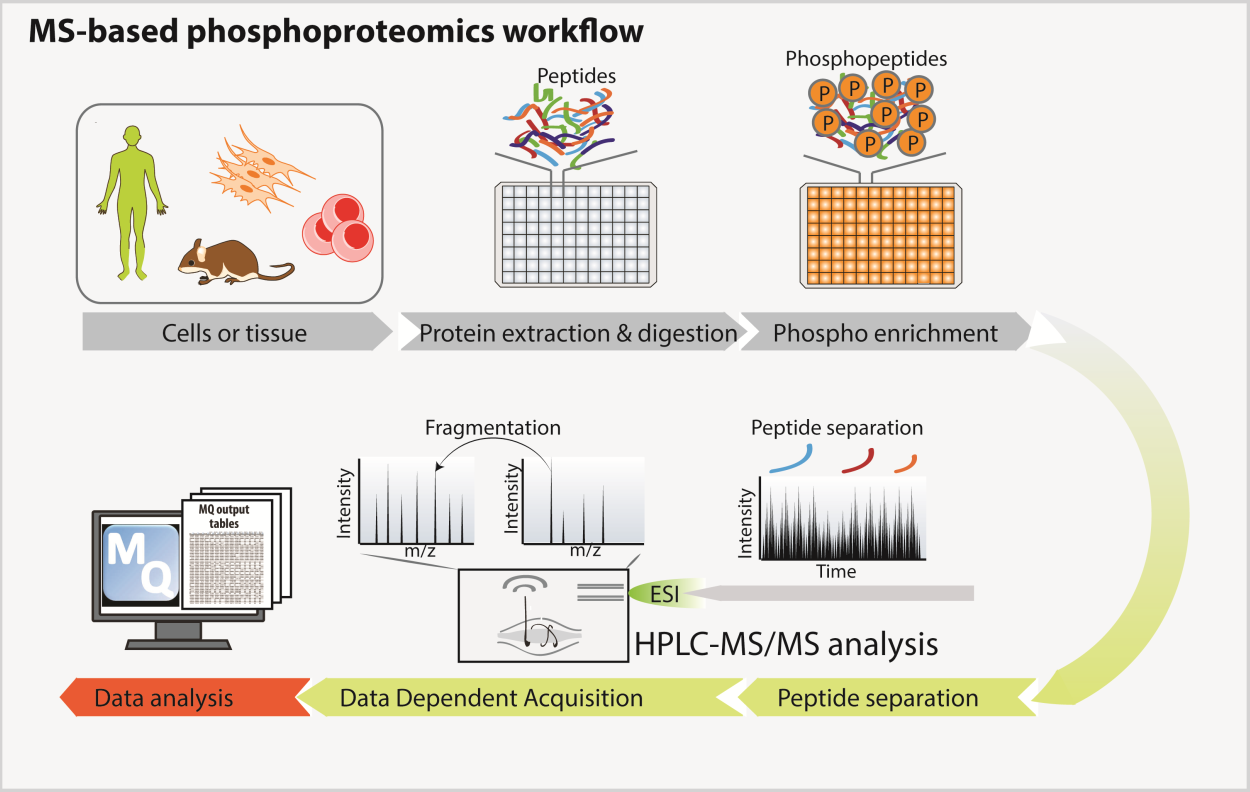
From Sacco et al, Proteomics, 2018
MS-based immuno-precipitation
Co-Immuno-precipitation experiments coupled with MS analysis are a potent tool to characterize protein complexes and their response to regulatory stimuli. In conjunction with specific bait molecules, proteins can be analysed in this way to provide quantitative information on protein binding partners and stoichiometry.
MS-based Co-IP workflow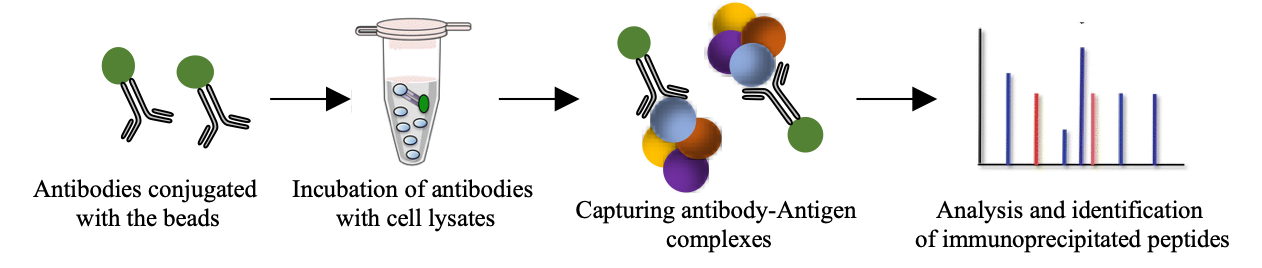
Instruments for LC MS/MS analysis consist of a Nano LC1200 coupled via a nano-electrospray ionization source to the quadrupole-based Q Exactive HF benchtop mass spectrometer. The generated spectral data are elaborated using Max Quant (MPI Martinsried), implemented using the Andromeda search engine, in order to obtain the list of identified proteins and their abundance in samples. For further analysis, the Perseus software is employed, and data are first filtered for contaminants and reverse entries as well as proteins that were only identified by a modified peptide.
