Assistant Investigator
Other positions:
Research Fellow, DICMaPI - Department of Chemical, Materials and Production Engineering, University of Naples "Federico II", Italy
Gennaro Gambardella received his M.Sc. Degree in Computer Science in November 2009 from the University of Naples "Federico II". In 2013, he completed his Ph.D. studies in “Bioinformatics and Computational Biology” at the University of Naples "Federico II", in the laboratory of Prof. Diego di Bernardo at Telethon Institute of Genetics and Medicine (TIGEM). He then held a position as a postdoctoral researcher at the King’s College of London (UK) in the laboratory of Dr. Francesca Ciccarelli until August 2015. From September 2015 to April 2018, he was a postdoctoral researcher in the laboratory of Prof. Diego di Bernardo at TIGEM. In May 2018, thanks to a 2-year STAR (Sostegno Territoriale alle Attività di Ricerca) grant, he became a junior Principal Investigator (PI) at the University of Naples "Federico II". In September 2020, he became an independent researcher at TIGEM in Naples where he now runs his own lab. From May 2020, he became Research Associate in the Department of Chemical, Materials and Production Engineering of the University of Naples “Federico II”. His research interests are strongly interdisciplinary, he works with a multidisciplinary team of biologists, mathematicians, oncologists, engineers and computer scientists who apply molecular genetics, data analysis and theoretical modeling to study cancer biology and other genetic diseases.
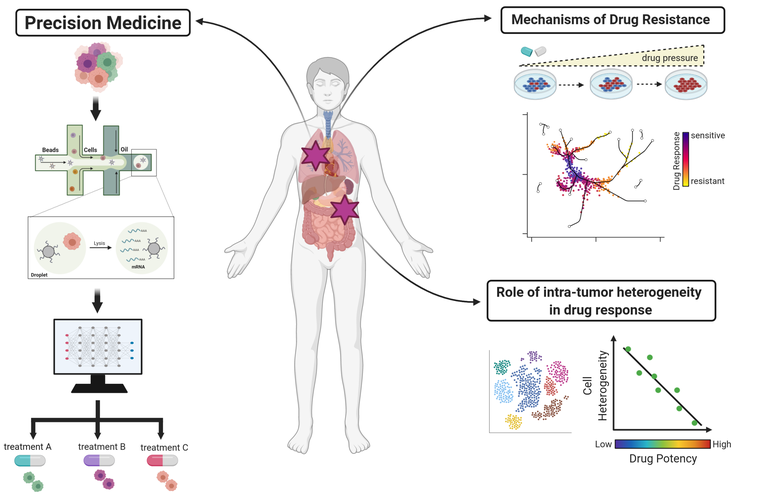
Our research goal is to understand the role and significance of genomic and non-genomic alterations that occur in cancer diseases (in particular Breast cancer and Lynch Disease) by using systems biology approaches.
Tumours are complex ecosystems characterized by a high degree of genomic, epigenomic and transcriptional variability (i.e., intra-tumour heterogeneity). Thus, therapies targeting early clonal alterations present at all
sites of the disease often have only short-term effects and drives the selection of resistant, rare and more aggressive sub-populations of cancer cells.
We therefore use system biology approaches to identify novel biomarker genes implicated in drug responses. This approach is extremely useful as it allows an individual patient’s genomic disease signature to be effectively translated into the most effective therapeutic intervention to apply. For example, we have shown that machine learning can be used to detect expression-based biomarker genes of drug sensitivity. This information can be used to predict from single-cell transcriptomic data which combination of compounds should be used to kill co-existing heterogenous sub-populations of breast cancer cells.
While we continue to characterize novel drug response biomarker genes, we also use them to (1) elucidate mechanisms of drug resistance; (2) assess the role of tumour heterogeneity on drug response and (3) to develop novel computational methods in the framework of precision medicine.
- GADD34 is a modulator of autophagy during starvation. Science Advances, 2020
- The impact of microRNAs on transcriptional heterogeneity and gene co-expression across single embryonic stem cells. Nature Communications, 2017
- A Tool for Visualization and Analysis of Single-Cell RNA-Seq Data Based on Text Mining. Frontiers in Genetics, 2019
- Patients with genetically heterogeneous synchronous colorectal cancer carry rare damaging germline mutations in immune-related genes. Nature Communications, 2016
- Single-Cell RNA Sequencing Analysis: A Step-by-Step Overview. Methods in Molecular Biology Book series, 2021
Complete List of Published Work in MyBibliography
Quote
Computational machine learning combined with human decision making, I believe, is the road we have to pursue for effective and individualized treatment.
Additional Funding
- Explore therapeutic resistance of triple negative breast cancer with single-cell transcriptomics and lineage tracing (2020-2024), AIRC






